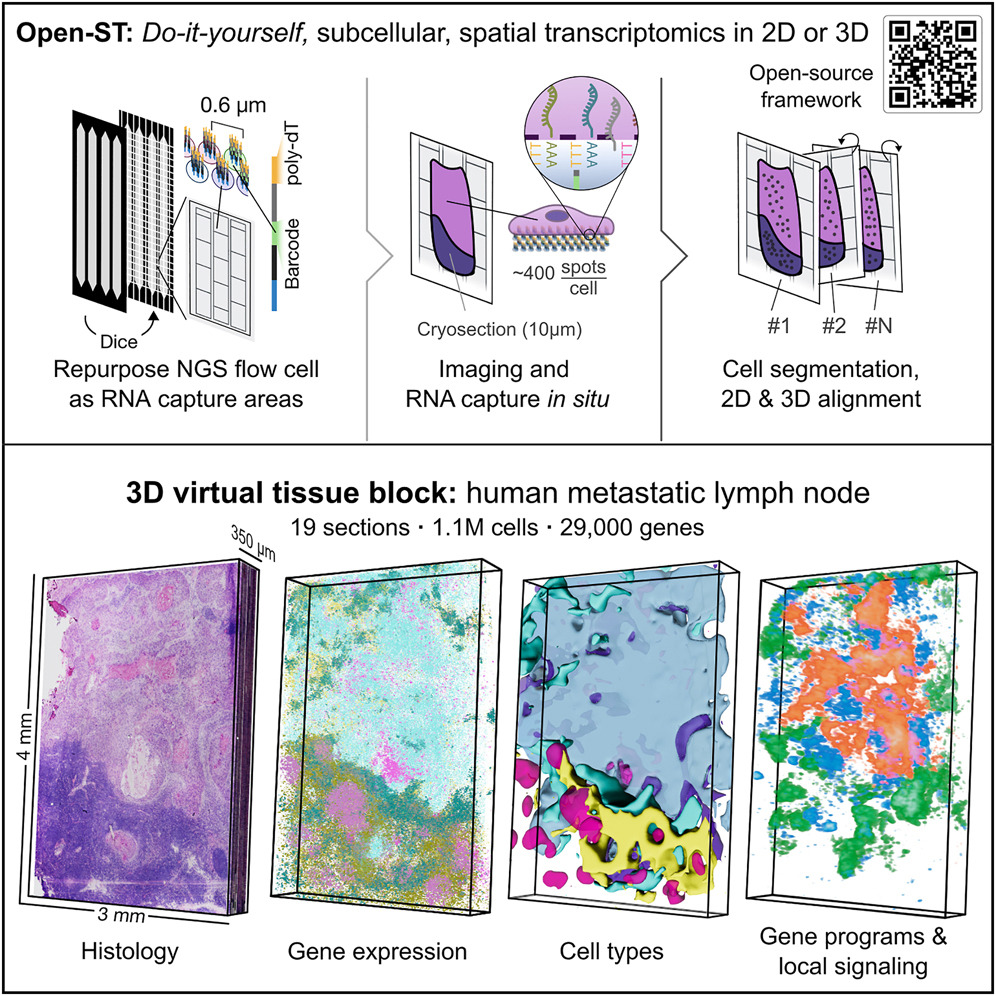
Until recently, performing detailed molecular analyses was not possible as available techniques could only obtain average values referred to a large number of molecules. Not only does Open-ST allow researchers to study the aspects of each cell and therefore their specific molecular contribution within a sample using “single-cell omics”, but it also allows researchers to define the spatial localization of each cell within a tissue.
Previously available ST technology could characterize molecules transcribed from each gene while preserving their localization but was limited due to high costs and poor resolution regarding the sensitivity in defining molecules in each single cell. As a result, the protocols behind Open-ST contain a precise sequence of experimental steps, including the analysis of molecular markers, the division of the cell into subunits, and finally the editing and digital visualization of the data through specially developed software.
Demonstrating its validity and efficiency, the international team tested cells taken from various types of tissues. Results from healthy and metastatic lymph nodes from patients with head and neck cancer correctly analyzed the high variability of the samples’ genetic code. The Open-ST approach can help scientists identify biomarkers, which is useful for the characterization of the tumor tissue itself.